Table des matières
Bench1
Description
- Type: Molecular dynamic
- Number of atoms: 80814
- Step size: 2fs
- Number of steps: 1e5
- Constraints:
- Ensemble: NPT (Langevin temperature: 310K, Langevin pressure: 1 atm)
- Rigid bonds (all)
- Electrostatics: Particle-Mesh Ewald
- Non bonded cutoff: 12 Angstroms
Results
- nodes: Number of nodes
- ntasks: Number of MPI tasks/node
- nthreads: Number of thread/task
- ngpus: Number of GPUs/node
- Wallclock: The time taken by the run (seconds)
- ns/day: The performance of the run (ns simulated/day of run)
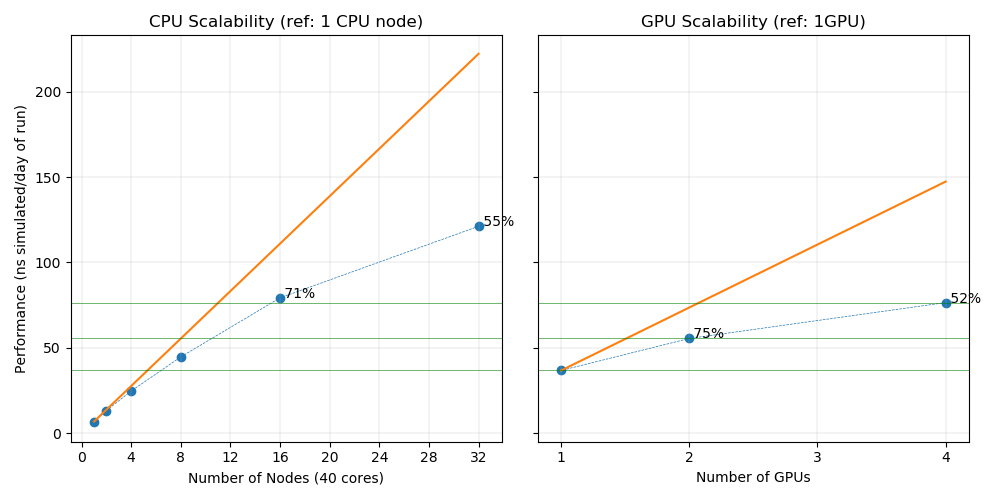
| nodes | ntasks | nthreads | ngpus | Wallclock (s) | ns/day | Parallel efficiency |
|---|---|---|---|---|---|---|
| 1 | 1 | 40 | 4 | 289.206665 | 76.55 | 52% (/1GPU) |
| 1 | 1 | 20 | 2 | 351.347107 | 55.55 | 75% (/1GPU) |
| 1 | 1 | 10 | 1 | 492.106934 | 36.83 | GPU reference |
| 32 | 40 | 1 | 0 | 168.47998 | 121.16 | 54% (/1CPU node) |
| 16 | 40 | 1 | 0 | 236.20047 | 79.32 | 71% (/1CPU node) |
| 8 | 40 | 1 | 0 | 403.368469 | 44.77 | 81% (/1CPU node) |
| 4 | 40 | 1 | 0 | 725.162048 | 24.84 | 89% (/1CPU node) |
| 2 | 40 | 1 | 0 | 1334.130371 | 13.35 | 96% (/1CPU node) |
| 1 | 40 | 1 | 0 | 2529.371826 | 6.94 | CPU reference |
Analysis
For this case the performance of 1 GPU is equivalent to 6 CPU nodes (240 cores). The parallel efficiency inside one GPU node is not good, dropping to 52% for 4 GPUs. This can be explained by the size of the system which is not sufficient to fill 4 GPUs.
The parallel efficiency of the CPU nodes drops quickly for similar reasons. In this case the most efficient way to use the GPU node is to run 4 simultaneous molecular dynamics.